Single-cell Genomics Assays
A curated list for interesting SCG methods
CITE-seq / Cell Hashing / REAP-seq
The CITE-seq / Cell Hashing / REAP-seq share the same philosophy in which a barcoded DNA oligo is conjugated with certain antibody, and during the scRNA-seq library preparation the oligo is captured by the reverse transcription primers as part of the cDNA library. By sequencing the antibody oligo library together with the standard 10X GEX library, one can simultaneously acquire the protein expression level and transcriptome from the same cell. If some universially expressed protein such as β2m is used, the method can also be used to label and multiplex samples from different conditions. And that’s where the variant method, Cell Hashing, is used for.
Resources
- Link to the CITE-seq Paper
- Link to the REAP-seq Paper
- Link to the Cell Hashing Paper
- CITE-seq / Cell Hashing Website
- Cell Hashing Protocol
Features
Pros
- Compatible with a wide variety of methods, including any poly-dT based scRNA-seq pipeline and 10X feature barcoding / 5’scRNA-seq workflow.
- Ease of doing.
- Antibody staining is very stable, and can survive FACS sorting of the cells.
- Commercially available through BioLegend.
Cons
- Price is relatively high (~$20 per staining) compared with MULTI-seq; but more affordable than 10X Cellplex.
- In situ labeling is yet to be tested, have to dissociate the samples into single cell suspension first.
- Limited numbers of barcodes commercially available (only 15).
- Customization of barcode is not easy (need customized antibody conjugation).
Variants of the method
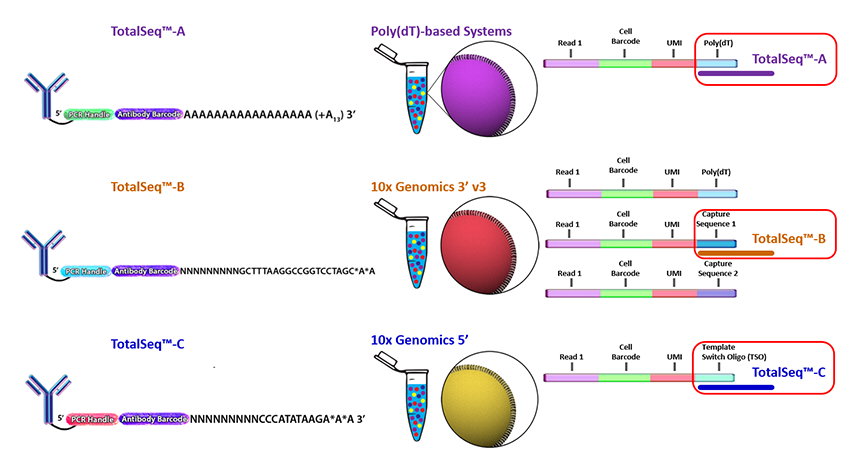
| Antibodies | 10X Compatibility | Captured Mechanism |
|---|---|---|
| TotalSeq-A | 3’scRNA-seq | dT Oligo on 10X beads |
| TotalSeq-B | 3’scRNA-seq with Feature Barcoding | CS1 |
| TotalSeq-C | 5’scRNA-seq | ADT-specific primer |
Full Protocols from 10X
| 10X Genomics Kits |
|---|
| 3’scRNA-seq LT Kit |
| 3’scRNA-seq Std Kit |
| 3’scRNA-seq HT Kit |