Single-cell Genomics Assays
A curated list for interesting SCG methods
Single-cell Genomics Assays
About this page
My name is Shiqi (Russell) Xie, currently a scientist in Genentech working on single-cell genomics. As a personal note, I started this page to summarize all the single-cell genomics and relevant technologies. The field is moving very fast and we see new methods coming out publishing almost every single month. And therefore the goal here is not to create the most comprehensive list for all the published assays, but only focus on the methods that I am actively using or could potentially be useful for my future projects. I will also comment about their usages.
Disclaimer
This website is for scientific discussion only. All the views, comments are my own.
If you have any questions or suggestions, please contact me through my email: russellxie@gmail.com.
Overview of all the 10X-related Technolgies
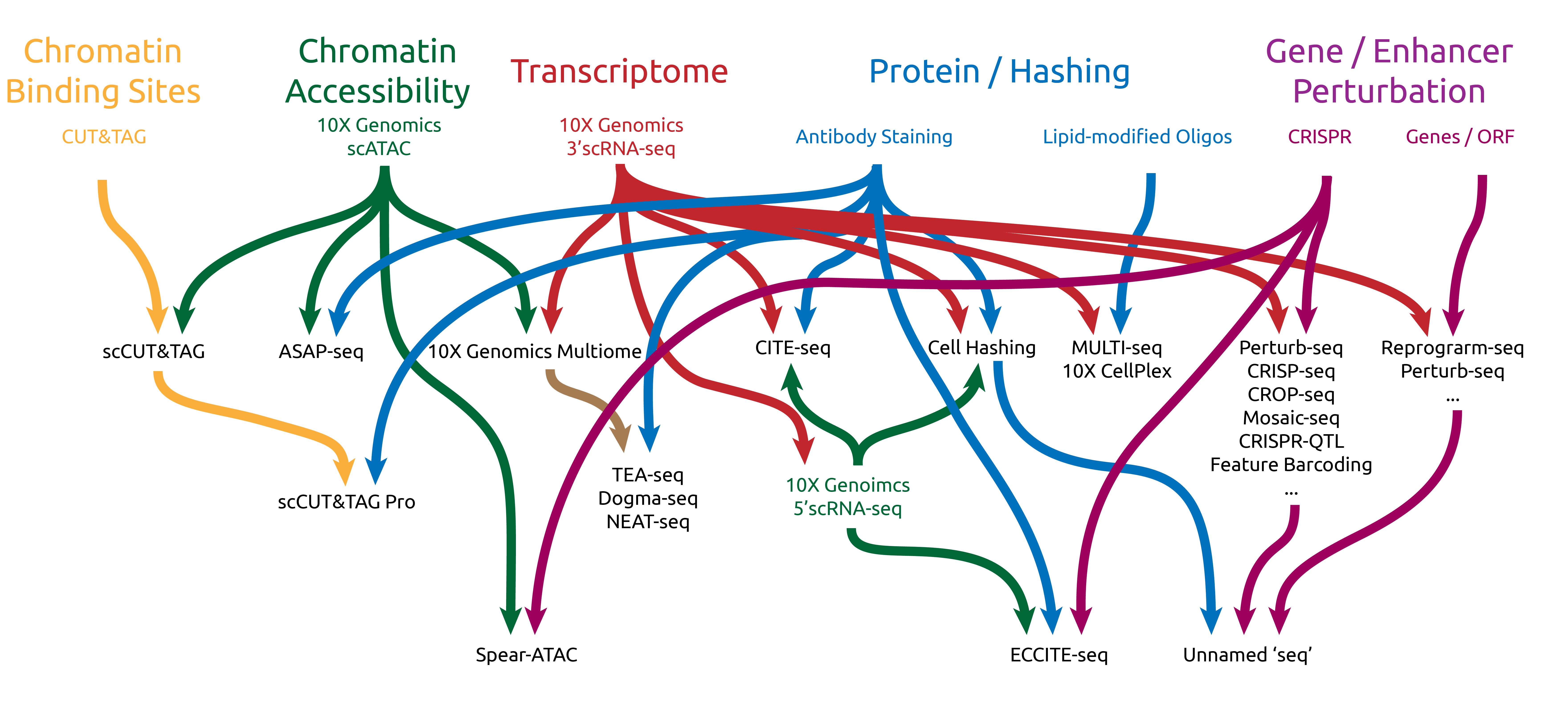
General Considerations
There are many directions that the field is making progress, including:
- Sample Multiplexing
- Higher Throughput
- Multiple Modality
- Easier sample preparation / storage
- Perturbations Introduction
- Lineage Tracing
Quick Comparison of Methods
Multi-modality / Multiplexing methods
| Method | Whole Cell | Nuclei | PFA Fixation | Methanol Fixation | Intracellular Protein | Modality |
|---|---|---|---|---|---|---|
| 10X Genomics 3’/5’ scRNA-seq | Yes | Yes | Coming soon* | Yes | No | RNA |
| MULTI-seq | Yes (LMO preferred) | Yes (CMO Preferred) | No | No | No | RNA |
| CellPlex | Yes | Yes | No | No | No | RNA |
| Cell Hashing / CITE-seq / REAP-seq | Yes | Yes | Coming soon* | NA | Coming Soon* | RNA + Protein |
| ASAP-seq | NA | Yes (Permeabilized Cell) | Yes (1%) | NA | Yes | ATAC + Protein |
| scCUT&TAG | NA | Yes | No | No | No | Chromatin Binding |
| scCUT&TAG-pro | NA | Yes | Yes (0.1%) | NA | Yes | Chromatin Binding + Protein |
| Dogma-seq / TEA-seq | NA | Yes (Permeabilized Cell) | No | NA | NA | RNA + ATAC + Protein |
| NEAT-seq | NA | Yes (Permeabilized Cell) | Yes (1.6%) | NA | Yes | RNA + ATAC + Protein |
*coming soon: 10X Genomics has annouced this feature earlier but the product has not been released.
Cost comparison of the multiplexing methods
| Method | Cost per reaction | Barcodes number | 10X compatibility |
|---|---|---|---|
| Cell Hashing / CITE-seq | $20 | 15* | 3’/5’ scRNA-seq |
| MULTI-seq | $0.2 | unlimited | 3’ scRNA-seq |
| CellPlex | $60 | 12 | 3’ scRNA-seq with CS2 |
* BioLegend sells cell hashing antibodies with 15 different barcodes on their website. It is possible one can get more than 15 by customized antibody conjugation or through a customized order from BioLegend.
Perturbation detection
| Method | Perturbation Method | Captured Feature | Barcode used | Modified lentivirus backbone | Capturing Method | 10X Compatibility |
|---|---|---|---|---|---|---|
| Perturb-seq | CRISPR | mRNA | Yes | Yes | dT oligo | 3’ scRNA-seq |
| CRISPR-seq | CRISPR | mRNA | Yes | Yes | dT oligo | 3’ scRNA-seq |
| Mosaic-seq | CRISPR | mRNA | Yes | Yes | dT oligo | 3’ scRNA-seq |
| CROP-seq | CRISPR | mRNA | No | Yes | dT oligoo | 3’ scRNA-seq |
| Feature Barcoding | CRISPR | sgRNA | No | Yes | Capture Sequence | 3’ scRNA-seq |
| ECCITE-seq | CRISPR | sgRNA | No | No | sgRNA-specific primer | 5’ scRNA-seq |
| Spear-ATAC | CRISPR | sgRNA | No | Yes | sgRNA-specific primer | scATAC-seq |
| Reprogram-seq | ORF Overexpression | mRNA | Yes | Yes | dT oligo / Capture Sequence | 3’ scRNA-seq |
Methods
Sample Multiplexing
- Cell Hashing (Antibody)
- MULTI-seq (Lipid-modified oligo, double stranded)
- 10X Genomics CellPlex (lipid-modified oligo, single-stranded)
- Genetic Multiplexing (computational method)
Higher Throughput
Split-and-pool methods
- sci-RNA-seq3
- scifi-RNA-seq
- SPLiT-seq
- 10X Genomics 3’ RNA-seq HT kit
Perturbations
CRISPR-based Methods
- Perturb-seq, Mosaic-seq, CRISP-seq (use barcodes to represent sgRNA)
- CROP-seq (detect mRNA to represent sgRNA)
- 10X Genomics Feature Barcoding (directly capture of sgRNA)
Overexpress ORFs
- Reprogram-seq
- Perturb-seq (a variant)
Lineage Tracing
- TraCe-seq
- Method through mtDNA
- scGESTALT
Multiple Modality
RNA + Protein
ATAC + Protein
- ASAP-seq
ATAC + Perturbation
- Spear-ATAC
RNA + ATAC
- 10X Genomics Multiome
- SHARE-seq
Chromatin Binding + Protein
- scCUT&TAG-pro
RNA + ATAC + Protein
- Dogma-seq, TEA-seq (cell surface proteins)
- NEAT-seq (Intracellular protein staining)
RNA + Protein + Perturbations
- ECCITE-seq